D2O-probed Raman Microspectroscopy Distinguishes Metabolic Dynamics of Macromolecules in Organellar Anticancer-Drug Response
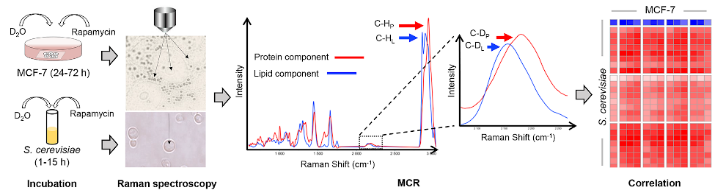
To profile the metabolic dynamics responding to drugs at the single-cell/organelle resolution, rapid and economical and mechanism-revealing methods are required. Here, we introduced D2O-probed Raman microspectroscopy in combination with the multivariate curve resolution-alternating least squares (MCRALS or MCR) algorithm. Exploiting MCR to deconvolute each macromolecular component specifically,the method is able to track and distinguish changes in lipid and protein metabolic activities in a human cancer cell line (MCF-7) and in Saccharomyces cerevisiae, in response to the metabolism-inhibitory effect of rapamycin. Both protein and lipid metabolic activities are suppressed after rapamycin treatment in the lipid bodies of cancer cells; however, in the nucleus, protein synthesis remains active, whereas lipid synthesis is inhibited. Thus, rapamycin differentially influences protein and lipid synthesis in mTOR signaling. Moreover, the strong correlation between macromolecular-specific components of yeast and those in MCF-7 cytoplasm, nucleus and lipid bodies revealed similarity in rapamycin response. Notably,highly metabolically active cancer cells after high-dosage rapamycin exposure (50 or 500 × IC50) were revealed, which escape detection by population-level cytotoxicity tests. Thus, by unveiling macromolecule-specific metabolic dynamics at the organelle level, the method is valuable to mechanismbased rapid screening and dissection of drug response.
| SCC Project ID | P_SCRS0201 |
| Project Relevance | Anticancer-drug response |
| Ecosystem | Host-associated, Engineered |
| SCC Organism Name | MCF7 ATCC HTB_22, Saccharomyces cerevisiae BY4742 |
| Add Date | 2020/10/21 |
| Add By | Maryam Hekmat Ara |
| PI | Jian Xu |
| Publication | NA |
| Number of Raman Spectra |